New L2R-2023:¶
More information about the new tasks: Here
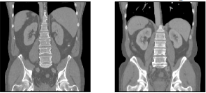
ThoraxCBCT (new)¶
ThoraxCBCT: Download here!

Citations, Data-Usage
Hugo, G. D., Weiss, E., Sleeman, W. C., Balik, S., Keall, P. J., Lu, J., & Williamson, J. F. (2016). Data from 4D Lung Imaging of NSCLC Patients (Version 2) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2016.ELN8YGLE
Hugo, G. D., Weiss, E., Sleeman, W. C., Balik, S., Keall, P. J., Lu, J., & Williamson, J. F. (2017). A longitudinal four-dimensional computed tomography and cone beam computed tomography dataset for image-guided radiation therapy research in lung cancer. In Medical Physics (Vol. 44, Issue 2, pp. 762–771). Wiley. https://doi.org/10.1002/mp.12059
NLST (extended)¶
NLST2023: Download here!


Citations, Data-Usage
National Lung Screening Trial Research Team. (2013). Data from the National Lung Screening Trial (NLST) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.HMQ8-J677
National Lung Screening Trial Research Team; Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD (2011). Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. New England Journal of Medicine, 365(5), 395–409. https://doi.org/10.1056/nejmoa1102873
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. DOI: https://doi.org/10.1007/s10278-013-9622-7
L2R-Dataset (pre-2022)¶
Lung CT
|
OASIS
|
CuRIOUS
|
|
|---|---|---|---|
| Modalities | CT -> CT | MR T1w -> MR T1w | MR T1w/FLAIR -> US |
| Patient Domain |
Intra-Patient (exhale / inhale) |
Inter-Patient | Intra-Patient |
| Resolution |
192x192x208 | 160x192x224 | 256x256x288 |
| Voxel size |
1.75x1.25x1.75mm | 1x1x1mm | ~0.5x0.5x0.5mm |
| Cases (train/test) |
30 (20/10) | 455 (416/39) | 32(22/10) |
| Preprocessing | affine pre-align crop/pad/resample |
resample | |
| Annotations | 100 landmarks/case | 35 anatomical labels | 9-18 landmarks/case |
| Additional Data | lung masks | ||
| Challenges | partial visibility small datasets large deformations unsupervised registration |
small structures | multi-modal scans small datasets unsupervised registration |
Download |
|
|
|
|
Abdomen CT-CT
|
Abdomen MR-CT
|
Hippocampus MR
|
|---|---|---|---|
| Modalities | CT -> CT |
MR T1w -> CT | MR T1w -> MR T1w |
| Patient Domain |
Inter-Patient | Intra-Patient | Inter-Patient |
| Resolution |
192x160x256 | 192x160x192 | 64x64x64 |
| Voxel size |
2x2x2mm | 2x2x2mm | 1x1x1mm |
| Cases (train/test) |
50 (30/20) | 16 (8/8) | 394 (263/131) |
| Preprocessing | canonical affine pre-align crop/pad/resample |
canonical affine pre-align crop/pad/resample |
crop/pad/resample |
| Annotations | 13 anatomical labels | 4 anatomical labels (training) 9 anatomical labels (test) |
2 anatomical labels |
| Additional Data | 90 unpaired MR/CT scans ROI masks |
||
| Challenges | small datasets large deformations |
multi-modal scans few/noisy annotations large deformations missing correspondences |
small structures |
Download |
|
|
|
Abdomen CT-CT¶

Description: All scans were captured during portal venous contrast phase with variable volume sizes (512 × 512 × 53 ~ 512 × 512 × 368) and field of views (approx. 280 × 280 × 225 mm3 ~ 500 × 500 × 760 mm3). The in-plane resolution varies from 0.54 × 0.54 mm2 to 0.98 × 0.98 mm2, while the slice thickness ranged from 1.5 mm to 7.0 mm.
Number of cases:Training: 30, Test: 20
Annotation: Thirteen abdominal organs were considered regions of interest (ROI), including spleen, right kidney, left kidney, gall bladder, esophagus, liver, stomach, aorta, inferior vena cava, portal and splenic vein, pancreas, left adrenal gland, and right adrenal gland. The organ selection was essentially based on [Shimizu A, Ohno R, Ikegami T, Kobatake H, Nawano S, Smutek D. Segmentation of multiple organs in non-contrast 3D abdominal CT images. International Journal of Computer Assisted Radiology and Surgery. 2007;2:135–142.]. As suggested by a radiologist, the heart was excluded for lack of full appearance in the datasets, and instead the adrenal glands were included for clinical interest. These ROIs were manually labeled by two experienced undergraduate students with 6 months of training on anatomy identification and labeling, and then verified by a radiologist on a volumetric basis using the MIPAV software.
Pre-processing: Common pre-processing to same voxel resolutions and spatial dimensions as well as affine pre-registration will be provided to ease the use of learning-based algorithms for participants with little prior experience in image registration.
Citation: Xu, Zhoubing et al. Evaluation of six registration methods for the human abdomen on clinically acquired CT, IEEE Transactions on Biomedical Engineering, 63 (8), pages=1563-1572, 2016
Abdomen MR-CT¶


TCIA Subject IDs Training/Validation: TCGA-B8-5158 (0002), TCGA-B8-5545 (0004), TCGA-B8-5551 (0006), TCGA-BP-5006 (0008), TCGA-DD-A1EI (0010), TCGA-DD-A4NJ (0012), TCGA-G7-7502 (0014), TCGA-G7-A8LC (0016)
Size: 122 CT/MR scans (16 CT-MR scan pairs (8 Training, 8 Test) + 90 unpaired CT/MR scans)
Source: TCIA, BCV, CHAOS
Challenge: Multimodal registration. Learning from few/noisy annotations. Learning with domain gaps.
Annotation on training data: Manual and automatic segmentations of different organs.
Annotation on test data: Manual segmentations of different organs.
Citation/Licence: Readme TCIA, Readme BCV, Readme CHAOS
Hippocampus MR¶

Description: The dataset consisted of MRI acquired in 90 healthy adults and 105 adults with a non-affective psychotic disorder (56 schizophrenia, 32 schizoaffective disorder, and 17 schizophreniform disorder) taken from the Psychiatric Genotype/Phenotype Project data repository at Vanderbilt University Medical Center (Nashville, TN, USA). Patients were recruited from the Vanderbilt Psychotic Disorders Program and controls were recruited from the surrounding community. The MRI data will show parts of the brain covering the hippocampus formation. The algorithm targets the alignment of two neighboring small structures (hippocampus head and body) with high precision on mono-modal MRI images between different patients (new insights into learning based registration due to a large-scale dataset). All images were collected on a Philips Achieva scanner (Philips Healthcare, Inc., Best, The Netherlands). Structural images were acquired with a 3D T1-weighted MPRAGE sequence (TI/TR/TE, 860/8.0/3.7 ms; 170 sagittal slices; voxel size, 1.0 mm3). The data is provided by the Vanderbilt University Medical Center (VUMC) and part of the medical segmentation decathlon.
Number of cases: Training: 263, Test: 131
Annotation: We use anatomical segmentations (and deformation field statistics) to evaluate the registration. Manual tracing of the head, body, and tail of the hippocampus on images was completed following a previously published protocol [Pruessner, J. et al. Volumetry of hippocampus and amygdala with high- resolution MRI and three-dimensional analysis software: minimizing the discrepancies between laboratories. Cerebral cortex 10, 433–442 (2000).; Woolard, A. & Heckers, S. Anatomical and functional correlates of human hippocampal volume asymmetry. Psychiatry Research: Neuroimaging 201, 48–53 (2012).]. For the purposes of this dataset, the term hippocampus includes the hippocampus proper (CA1-4 and dentate gyrus) and parts of the subiculum, which together are more often termed the hippocampal formation [Amaral, D. & Witter, M. The three-dimensional organization of the hip- pocampal formation: a review of anatomical data. Neuroscience 31, 571– 591 (1989)]. The last slice of the head of the hippocampus was defined as the coronal slice containing the uncal apex.
Pre-Processing: Common pre-processing to same voxel resolutions and spatial dimensions will be provided to ease the use of learning-based algorithms for participants with little prior experience in image registration.
Citation: A Simpson et al.: "A large annotated medical image dataset for the development and evaluation of segmentation algorithms" arXiv 2019
Lung CT¶


Size: 30 3D volumes (20 Training + 10 Test)
Source: Department of Radiology at the Radboud University Medical Center, Nijmegen, The Netherlands.
Challenges: Estimating large breathing motion. The lungs are not fully visible in the expiration scans.
Annotation on training data: automatic lung segmentation + keypoints
Annotation on test data: manual landmarks
Citation: Hering, Alessa, Murphy, Keelin, & van Ginneken, Bram. (2020). Learn2Reg Challenge: CT Lung Registration - Training Data [Data set]. Zenodo. http://doi.org/10.5281/zenodo.3835682;
OASIS¶


Size: 416 3D MR scans
Source: OASIS dataset
Challenge: alignment of small structures of variable shape and size with high precision on mono-modal MRI images between different patients.
Annotation on training data: automatic segmentation processed with FreeSurfer and SAMSEG for the neurite package.
Annotation on test data: automatic segmentation processed with FreeSurfer and SAMSEG for the neurite package.
Citation: Open Access Series of Imaging Studies (OASIS):
Cross-Sectional MRI Data in Young, Middle Aged, Nondemented, and
Demented Older Adults.
Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL.
Journal of Cognitive Neuroscience, 19, 1498-1507.
HyperMorph: Amortized Hyperparameter Learning for Image
Registration.
Hoopes A, Hoffmann M, Fischl B, Guttag J, Dalca AV.
IPMI 2021.
Detailed Description: These data were prepared by Andrew Hoopes and Adrian V. Dalca for the following HyperMorph paper. If you use this collection please cite the following and refer to the OASIS Data Use Agreement. Evaluation for this challenge is performed on the images that are resampled into the affinely-aligned, common template space (Feel free to use any versions (aligned/non-aligned, raw/corrected) for tuning/training your algorithm)!
CuRIOUS¶
Description: The database contains 22 subjects with low-grade brain gliomas and is intended to help develop MRI vs. US registration algorithms to correct tissue shift in brain tumor resection. For the task, we included the T1w and T2-FLAIR MRIs, and spatially tracked intra-operative ultrasound volumes taken after craniotomy and before resection started. Matching anatomical landmarks were annotated between T2FLAIR MRI and 3D ultrasound volumes to help validate registration algorithm accuracy. All scans were acquired for routine clinical care of brain tumor resection procedures at St Olavs University Hospital (Trondheim, Norway). For each clinical case, the pre-operative 3T MRI includes Gadolinium-enhanced T1w and T2 FLAIR scans, and the intra-operative US volume was obtained to cover the entire tumor region after craniotomy but before dura opening, as well as after resection was completed. A detailed user manual is included in the data package download.
Number of cases: Training: 22, Test: 10
Annotations: Matching anatomical landmarks were annotated between T2FLAIR MRI and 3D ultrasound volumes. The reference anatomical landmarks were selected by Rater 1 in the ultrasound volume before dura is open after craniotomy. Then two raters selected matching anatomical landmarks in the ultrasound volumes obtained after tumor resection using the software 'register' from MINC Toolkit. The ultrasound landmark selection was repeated twice for each rater with a time interval of at least one week. Finally, the results (4 points for each landmark location) were averaged. Eligible anatomical landmarks include deep grooves and corners of sulci, convex points of gyri, and vanishing points of sulci. Same raters produced the anatomical landmarks for both the training and testing data. To help with deep learning purposes, the landmarks have also been voxelized as spheres in the same 3D space as the MRI/US scans. In addition, the image header transforms are also provided separately for the users.
Pre-Processing: All MRI scans were corrected for field inhomogeneity, and T1w MRI is rigidly registered to T2FLAIR MRI. For each subject, the MRI and 3D ultrasound volumes were resampled to the same space and dimension (256x256x288) at an isotropic ~0.5mm resolution.
Non-disclosure Agreement: Unlike the training data, which were publicly released as the EASY-RESECT database, the CuRIOUS 2020 for Learn2Reg Challenge test data are not public. They cannot be shared and used for any purpose but the challenge evaluation.
Citation: Y. Xiao, M. Fortin, G. Unsgård , H. Rivaz, and I. Reinertsen, “REtroSpective Evaluation of Cerebral Tumors (RESECT): a clinical database of pre-operative MRI and intra-operative ultrasound in low-grade glioma surgeries”. Medical Physics, Vol. 44(7), pp. 3875-3882, 2017.