Learn2Reg 2025¶
LUMIR25 Ranking¶
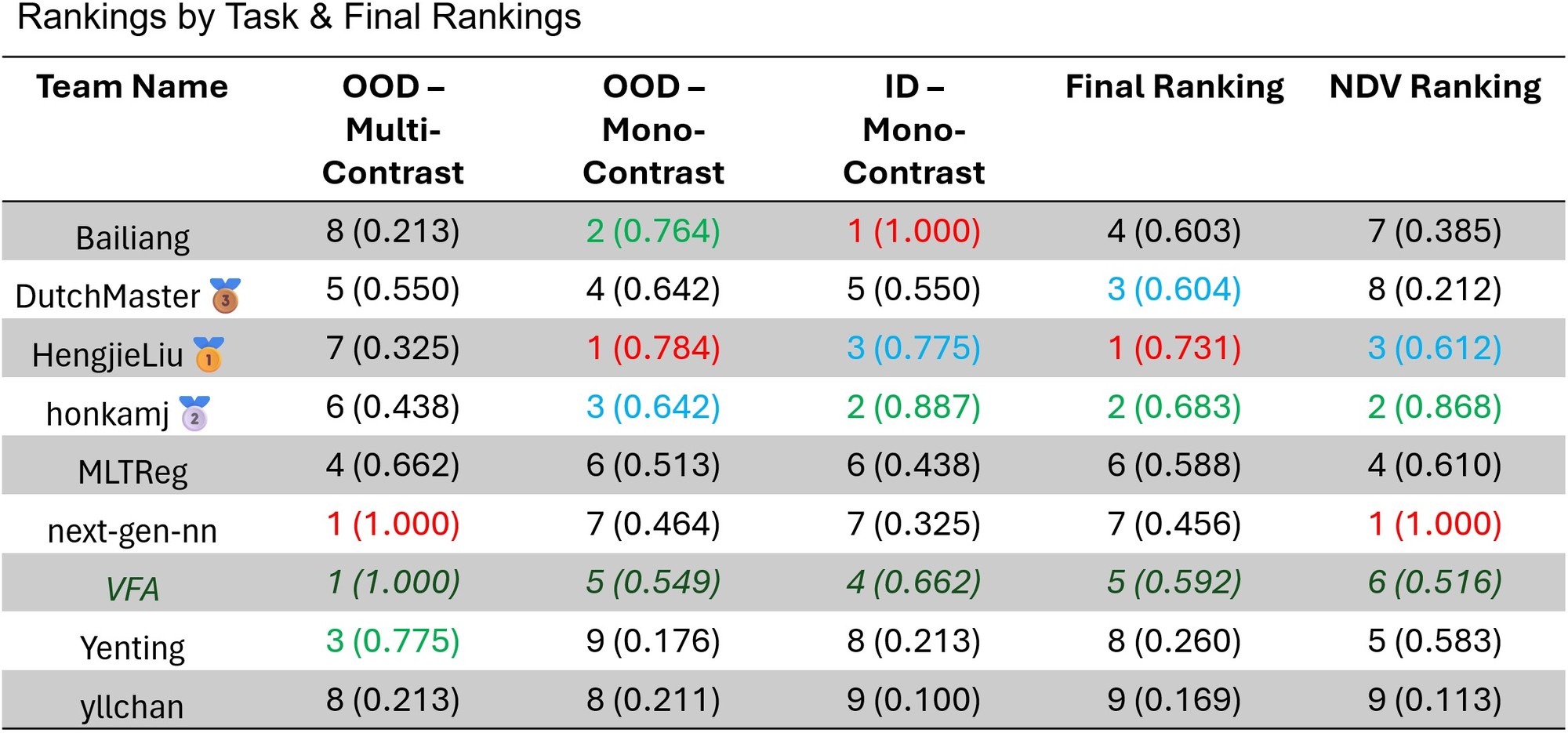 Detailed, per-dataset rankings and scores can be found here.
Detailed, per-dataset rankings and scores can be found here.
ReMIND2Reg Ranking¶
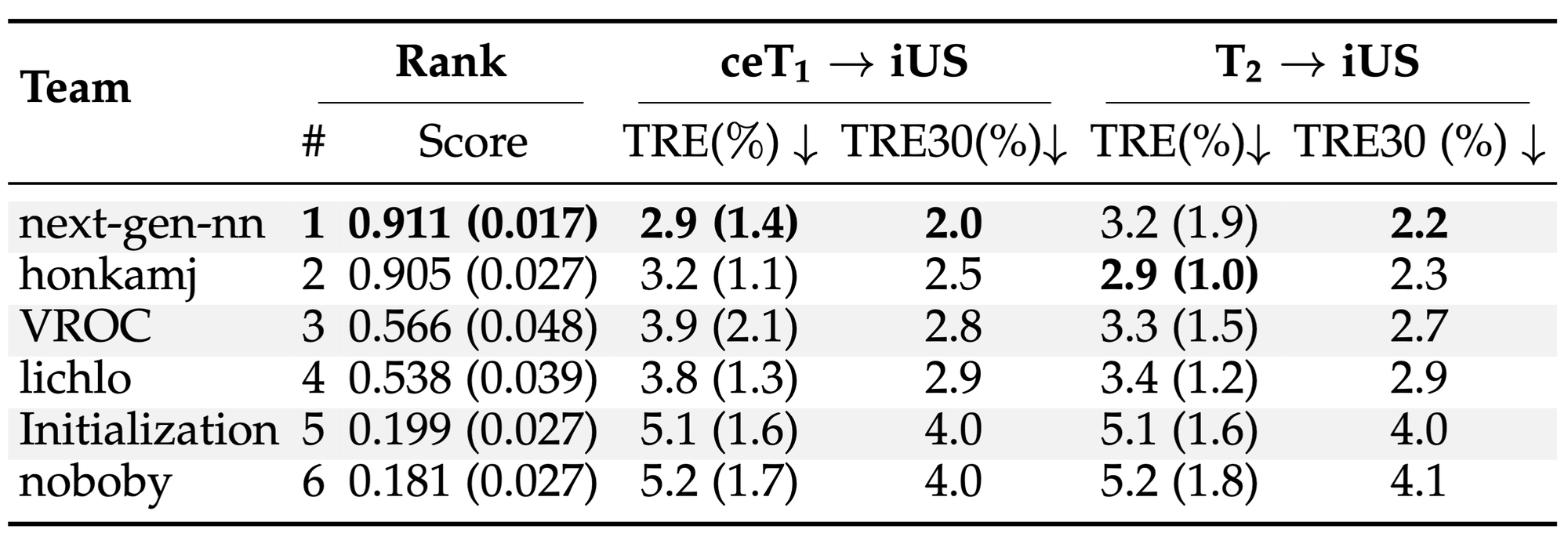 ¶
¶
We are pleased to host the Learn2Reg Challenge again this year at MICCAI 2025 in Daejeon!


If you like to be added to the Learn2Reg newsletter, please fill out this form.
In summary, Learn2Reg 2025 will feature two extended sub-tasks: LUMIR 2025 and ReMIND2Reg 2025. The full timeline is available here.
Important Dates:
- End of April: Release of training and validation datasets
- June 2nd: Kick-off meeting with live Q&A at 9:00 AM ET on Monday (Slides)
- Aug. 6th: Second virtual Q&A session at 10:00 AM ET on Wednesday (Zoom link)
- Aug. 8th (Tentative): Validation leaderboard ranking announced for early acceptance consideration
- End of August: Final submission deadline
This year, we are also offering the opportunity for invited participants to submit a full description of their methods to a special paper collection hosted by the MELBA journal.
In addition, prizes will be awarded thanks to generous sponsorship from SIGBIR, and each participant will receive a color-printed certificate:
- ReMIND25: 1st – $250, 2nd – $150, 3rd – $100
- LUMIR25: 1st – $250, 2nd – $150, 3rd – $100
Check this list out for answers to FAQs.
ReMIND 2025¶

(a) Contrast-enhanced T1 and post-resection intra-operative US; (b) T2 and post-resection intra-operative US.
Training/Validation: Download
Context: Surgical resection is the critical first step for treating most brain tumors, and the extent of resection is the major modifiable determinant of patient outcome. Neuronavigation has helped considerably in providing intraoperative guidance to surgeons, allowing them to visualize the location of their surgical instruments relative to the tumor and critical brain structures visible in preoperative MRI. However, the utility of neuronavigation decreases as surgery progresses due to brain shift, which is caused by brain deformation and tissue resection during surgery, leaving surgeons without guidance. To compensate for brain shift, we propose to perform image registration using 3D intraoperative ultrasound.
Objectives: The goal of the ReMIND2Reg challenge task is to register multi-parametric pre-operative MRI and intra-operative 3D ultrasound images. Specifically, we focus on the challenging problem of pre-operative to post-resection registration, requiring the estimation of large deformations and tissue resections. Preoperative MRI comprises two structural MRI sequences: contrast-enhanced T1-weighted (ceT1) and native T2-weighted (T2). However, not all sequences will be available for all cases. For this reason, developed methods must have the flexibility to leverage either ceT1 or T2 images at inference time. To tackle this challenging registration task, we provide a large non-annotated training set (N=158 pairs US/MR). Model development is performed on annotated validation sets (N=10 pairs US/MR). The final evaluation will be performed on a private test set using Docker (more details will be provided later). The task is to find one solution for the registration of two pairs of images per patient: 1. 3D post-resection iUS (fixed) and ceT1 (moving). 2. 3D post-resection iUS (fixed) and T2 (moving).
Dataset: The ReMIND2Reg dataset is a pre-processed subset of the ReMIND dataset, which contains pre- and intra-operative data collected on consecutive patients who were surgically treated with image-guided tumor resection between 2018 and 2024 at the Brigham and Women’s Hospital (Boston, USA). The training (N=99) and validation (N=5) cases correspond to a subset of the public version of the ReMIND dataset. Specifically, the training set includes images of 99 patients with 99 3D iUS, 93 ceT1, and 62 T2 and validation images of 5 patients with 5 3D US, 5 ceT1, and 5 T2. The images are paired as described above with one or two image pairs per patient, resulting in 155 image pairs for training and 10 image pairs for validation. The test cases are not publicly available and will remain private. For details on the image acquisition (scanner details, etc.), please see https://doi.org/10.1101/2023.09.14.23295596
Number of registration pairs: Training: 155, Validation: 10, Test: 40 (TBC).
Rules: Participants are allowed to use external datasets if they are publicly available. The authors should mention that these datasets were used in their method description, including references and links. However, participants are not allowed to use private datasets. Moreover, they cannot exploit manual annotations that were not made publicly available.
Pre-Processing: All images are converted to NIfTI. When more than one pre-operative MR sequence was available, ceT1 was affinely co-registered to the T2 using NiftyReg; Ultrasound images were resampled in the pre-operative MR space. Images were cropped in the field of view of the iUS in an image size of 256x256x256 with a spacing of 0.5x0.5x0.5mm.
Citation: P. Juvekar, et al., (2023). The Brain Resection Multimodal Imaging Database (ReMIND). Nature Scientific Data. https://doi.org/10.1101/2023.09.14.23295596
LUMIR 2025¶

GitHub: https://github.com/JHU-MedImage-Reg/LUMIR25_L2R25
Training: Download Validation: Download Simplified Validation Labels: Download Dataset JSON: Download
Note: The training set remains unchanged from LUMIR 2024.
Context: Last year’s LUMIR challenge highlighted the strong performance of deep learning-based methods, which outperformed traditional optimization-based approaches for mono-modal image registration under an unsupervised setting. These methods also exhibited notable robustness to domain shifts, maintaining high performance across datasets with varying image contrasts and resolutions.
For this year’s LUMIR challenge, we continue using the same training dataset, T1-weighted brain MRIs from healthy controls, but introduce more demanding validation scenarios. Participants are again asked to train their models on this dataset; however, the evaluation will additionally focus on zero-shot tasks that involve substantial domain shifts, such as high-field MRI, pathological brains, and alternate MRI contrasts.
In addition, we are expanding the zero-shot benchmark to include multi-modal registration tasks (e.g., T1- to T2-weighted MRIs, and potentially other modality pairs), offering a more comprehensive assessment of model generalizability across diverse neuroimaging applications.
To support participants, we also provide coarse anatomical label maps (10 classes) for the validation datasets, enabling participants to perform quick self-validation prior to leaderboard submission. However, evaluations on the leaderboard as well as the test phase will be conducted on much finer anatomical structures (over 100 classes).
Dataset: The released image data includes the OpenBHB dataset, featuring T1-weighted brain MRI scans collected from 10 publicly available datasets. A portion of the data also comes from the AFIDs project, based on the OASIS dataset. Consistent with the challenge's focus on unsupervised image registration, only imaging data will be provided for training, allowing participants to freely generate inter-subject pairs. All images have been converted to NIfTI format, resampled, and cropped to a standardized region of interest, resulting in a uniform image size of 160×224×192 voxels with 1×1×1 mm isotropic resolution. The validation dataset comprises:
- 10 in-domain subjects from the same distribution as the training set,
- 10 out-of-domain subjects with landmark annotations from a different dataset of healthy controls,
- 10 out-of-domain subjects from high-field MRI with anatomical segmentations (not included in training), and
- 10 multi-modal subjects for T1- to T2-weighted registration
The evaluation dataset includes the 590 test subjects from last year’s challenge, as well as an expanded zero-shot benchmark with over 1,000 previously unseen subjects from diverse sources. To ensure a true zero-shot evaluation, the zero-shot test set is entirely distinct from both the training and validation sets. Importantly, participants should NOT assume that the validation set fully represents the test scenarios. The dataset includes:
- Training images: 3,384 subjects
- Validation images: 40 subjects across four subgroups (as detailed above)
- Test images: 590 from the previous year + 1,000+ for zero-shot tasks
Dataset references include:
- Chen, Junyu, et al. "Beyond the LUMIR challenge: The pathway to foundational registration models." arXiv preprint arXiv:2505.24160 (2025).
- Dufumier, Benoit, et al. "Openbhb: a large-scale multi-site brain mri data-set for age prediction and debiasing." NeuroImage 263 (2022): 119637.
- Taha, Alaa, et al. "Magnetic resonance imaging datasets with anatomical fiducials for quality control and registration." Scientific Data 10.1 (2023): 449.
- Marcus, Daniel S., et al. "Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults." Journal of cognitive neuroscience 19.9 (2007): 1498-1507.
Frequently Asked Questions:¶
-
- Q: (General): Will snapshot validation rankings be based on the leaderboard ranking (i.e., Dice)?
- A: No. Snapshot rankings will be based on the average rank across all evaluation metrics. A method that performs well on Dice but poorly on other metrics is unlikely to rank highly overall.
-
- Q: (LUMIR25): Is the use of external datasets allowed?
- A: No. Only the dataset provided for the LUMIR challenge may be used.
-
- Q: (LUMIR25): Can I use image synthesis tools?
- A: Yes. You may use pre-existing, published image synthesis tools in any way you find effective. However, you may not train or fine-tune these tools using external data, as that would violate the restriction on using external datasets.
-
- Q: (LUMIR25): Will the same modalities appear in the final test phase?
- A: Not necessarily. The validation set may not fully represent the diversity of the final test cases. The test set may include additional brain imaging modalities, and their exact combinations will not be disclosed to preserve the zero-shot setting.
-
- Q: (LUMIR25): Can I use image segmentation tools?
- A: No. Image segmentation tools are not allowed. The LUMIR challenge focuses on unsupervised image registration. Using segmentation introduces bias from anatomical label maps, which can lead to unrealistic, non-smooth deformations.
The Microsoft CMT service was used for managing the peer-reviewing process for this conference. This service was provided for free by Microsoft and they bore all expenses, including costs for Azure cloud services as well as for software development and support.
